We translate biological interactions into numbers and equations to deduce new insights.
In systems biology we translate existing biological knowledge into numbers and equations. In this way we use computer simulations and the rules of mathematics to gain new insight to make better decisions across a wide range of applications – e.g., patient treatment, food production, and natural resource management.
Model-based phenotypes from omics data
Biological data is now being produced in such quantity that it can only be interpreted with the help of computational modeling; encoding, revising and improving our mental models of how biological systems work. The value of modeling is that it simplifies reality, allowing us to generalize insights from the model to a broader class of systems. Thus, zebrafish might represent fish in general, a slice of liver in a lab dish can represent the intact liver, and a set of differential equations can approximate the dynamic behavior of a biological system. Mathematical analysis can reveal hidden connections in existing data, as well as our own underlying assumptions, allowing us to deduce new insights or disprove false hypotheses.
An example of a generalized model is this simplified four-legged animal:
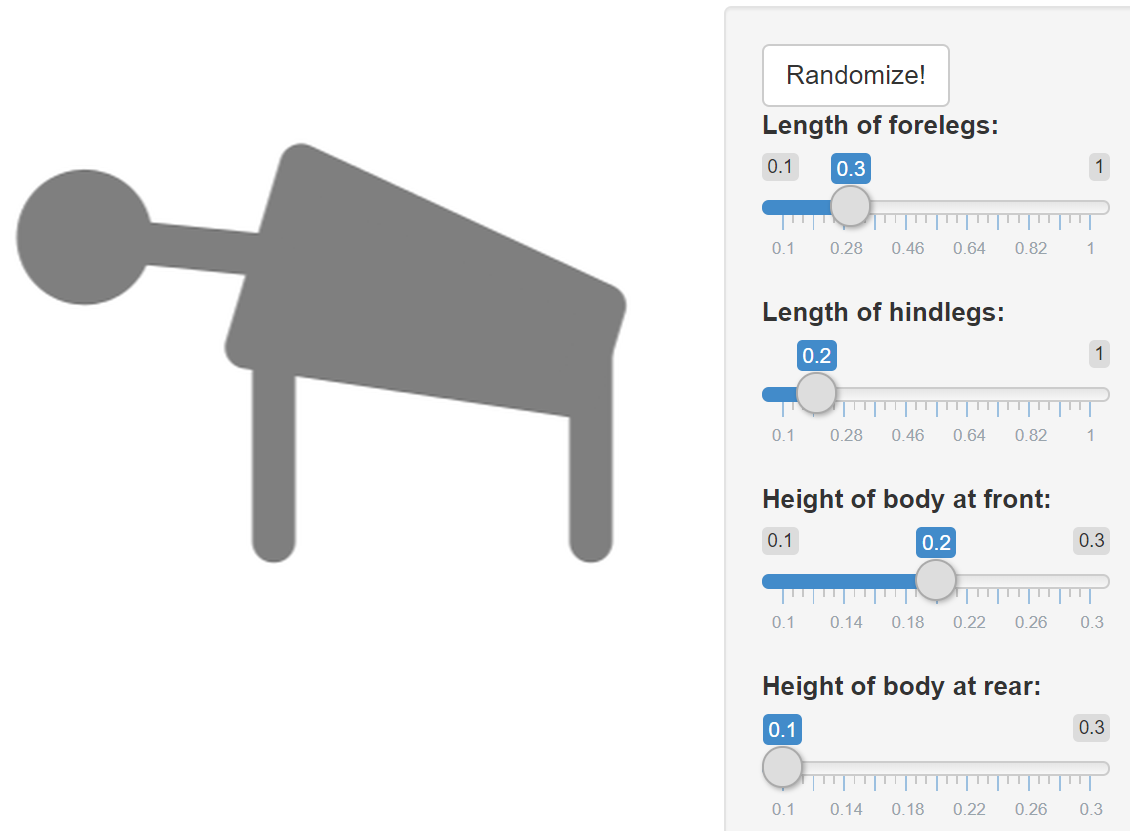
Modeling a class of systems allows us to see what these systems have in common (the mathematical form of the equations) and how they differ (in their respective parameter values). The parameters we look at are model-based phenotypes that condense vast numerical phenotypic data into fewer, more informative measurements. This phenotypic variation can then be traced back to genotypic variation – sometimes very directly, as with enzymes in metabolic networks, or ion channels in heart or nerve cells, but more often in pragmatic combination with statistical genetics and multivariate analysis.
Digital production biology: the Digital Salmon
The Digital Salmon project spearheads the digitization of data and knowledge about sustainable and efficient food production. It is part of Digital Life Norway, the Research Council's largest program for systems biology. The program applies mathematics to life, technology and value generation. With DigiSal we aim to explore the interactions between genetics and feed ingredients, in order to create more sustainable salmon production.
Watch the following video to learn more about DigiSal:
<strong>Click here for further reading:</strong>
Facts
Key Collaborators
- Daniel Kling, AquaGen, salmon breeding company
- Centre for Digital Life Norway
- NTNU Biotechnology
- Systems and Synthetic Biology, Wageningen University and Research Centre, Netherlands
